Genetics 1G
Mutation - some examples
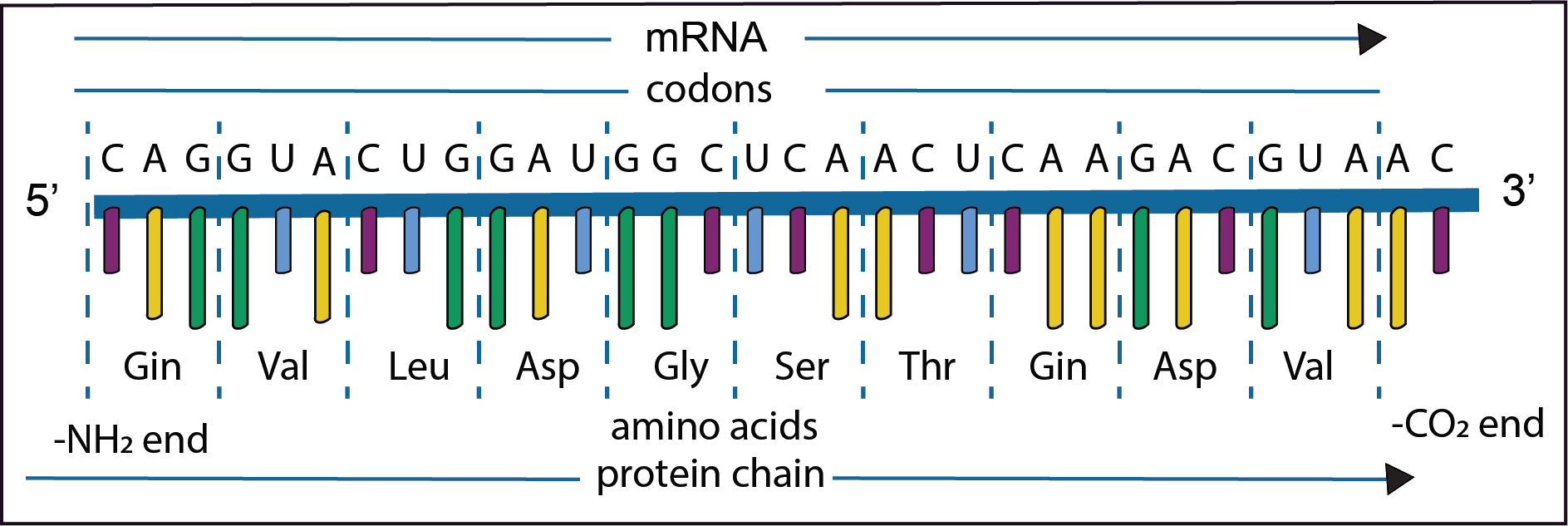
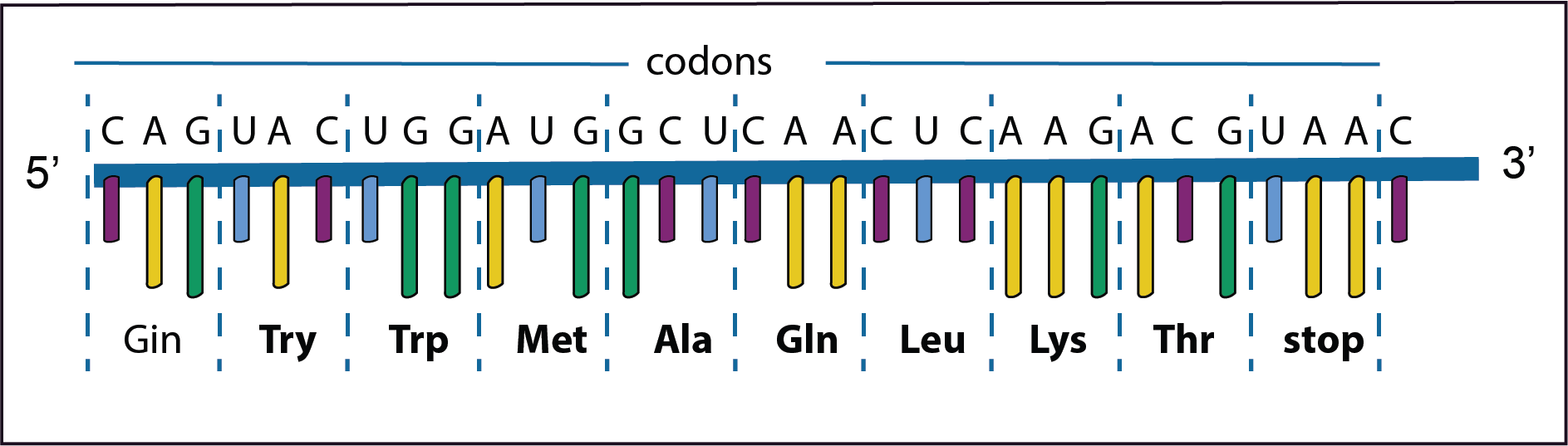
The diagram represents only a short part of the second step in the gene expression in which the information on the newly synthesised mRNA is used to direct the synthesis of a polypeptide. The order of the bases on the newly formed mRNA is shown in the diagram together with the codons and their respective amino acids. The mRNA is synthesised from left to right in the 5’ to 3’ direction. As has already been noted DNA replication is for the most part extremely accurate,
However, mistakes do occur resulting in mutations; changes in the nucleotide sequence in the mRNA. Some types are illustrated in the diagrams below. Text in bold indicates where a change has taken place.
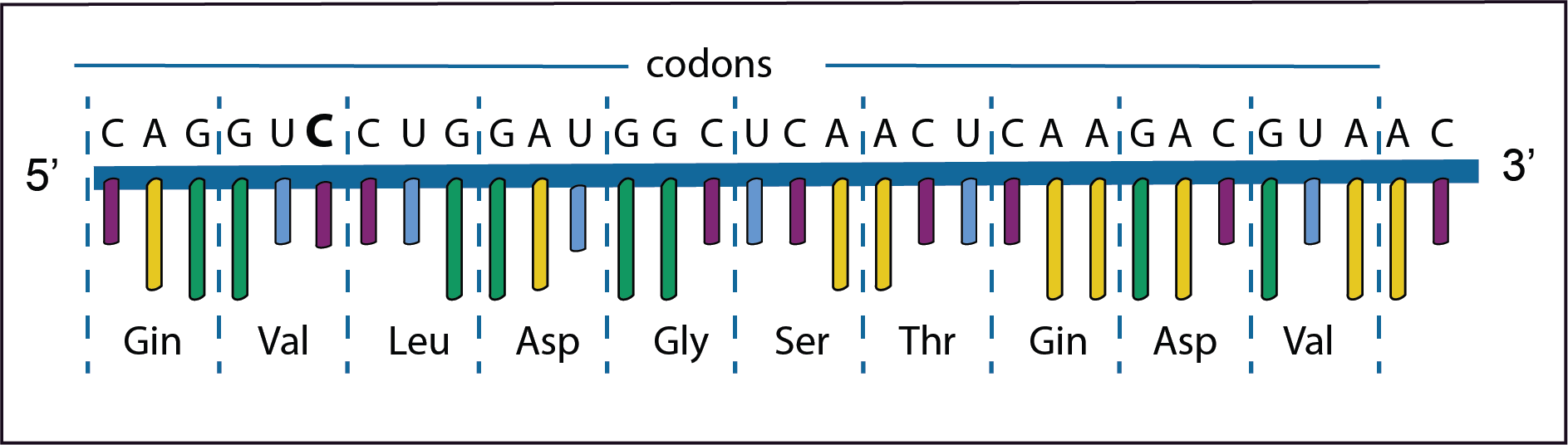
In the first example the third base A in the second codon has changed to C. However, there is no change in the amino acid Valine because GUA and GUC encode the same amino acid; this is called a synonymous mutation. Such a mutation has no effect on the protein’s structure.
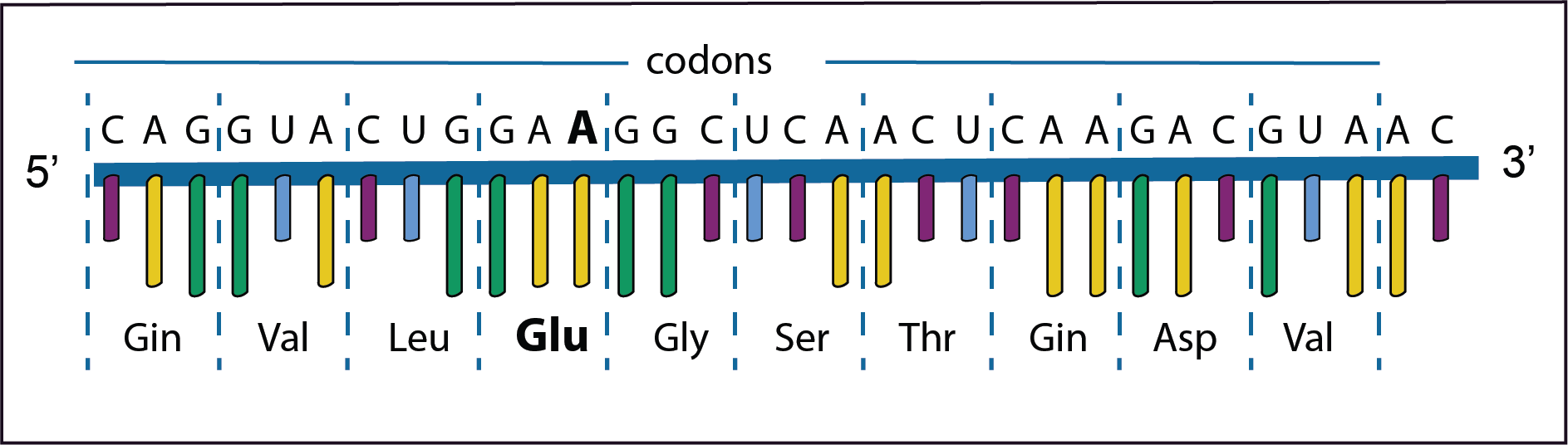
In this example in the forth codon GAU changes to GAA with a consequence that the amino acid changes from Asp to Glu.
This is called a missense mutation. The polypeptide with such an amino acid change may result in a changed phenotype depending if the changed amino acid plays a role in the structure or function of the protein.
This is called a missense mutation. The polypeptide with such an amino acid change may result in a changed phenotype depending if the changed amino acid plays a role in the structure or function of the protein.
Frameshift mutations occur if there is an addition or a deletion of a base. This results in a complete new set of amino acids being formed.
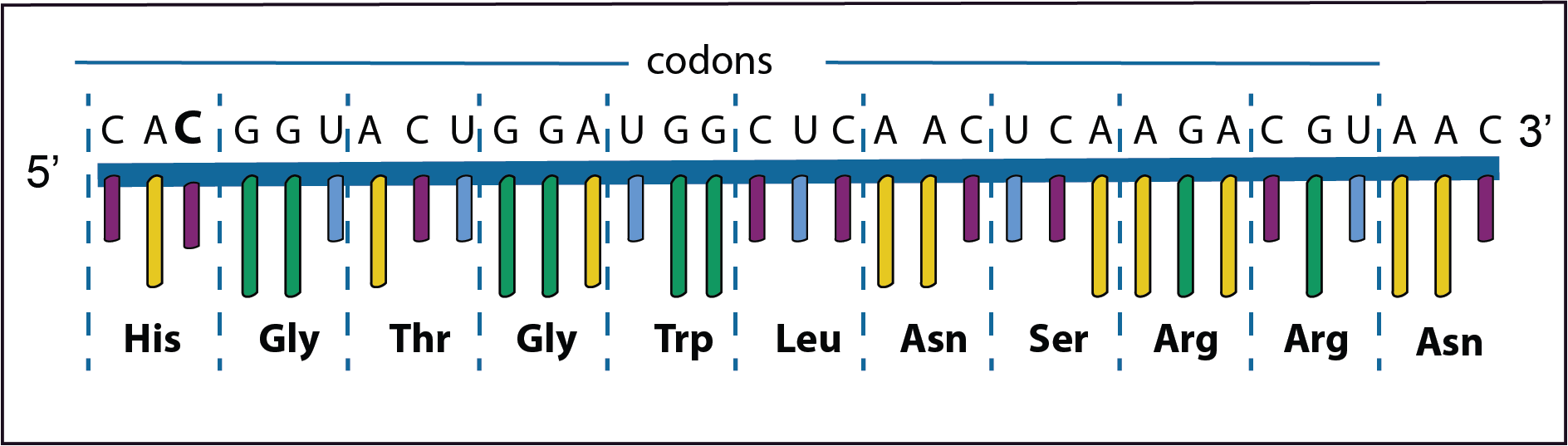
In the example on the left the third base (C) in the first codon has been added. This results in a frameshift of the codons to the left as such a new set of amino acids are formed.
In the example on the left the third base (C) in the first codon has been added. This results in a frameshift of the codons to the left as such a new set of amino acids are formed.
In the second example the original third base (G) has been deleted. Now there is a frameshift to the right again resulting in a new set of amino acids. Coincidentally the last codon is a stop codon so that the synthesis of the protein is halted prematurely. This is called a nonsense mutation.
In these two example the frameshift may result in mutant phenotypes.
In these two example the frameshift may result in mutant phenotypes.
Some mutations can be harmful or even lethal in which case natural selection will remove them from the population.
A small fraction of mutations are beneficial, improving the organism’s fitness. These are extremely important for evolution, since their directional selection leads to adaptive evolution.
When a mutation occurs in a gene it produces an allele. When different alleles are present in a bee’s population the population is said to be polymorphic. As seen in the examples above these alleles may encode slightly differnt versions of a protein giving rise to different phenotypic traits. A gene’s most common allele is call the wild type with rare or uncommon alleles called mutants. Natural selection and genetic drift determine the relative frequencies of the alleles in a population.
A small fraction of mutations are beneficial, improving the organism’s fitness. These are extremely important for evolution, since their directional selection leads to adaptive evolution.
When a mutation occurs in a gene it produces an allele. When different alleles are present in a bee’s population the population is said to be polymorphic. As seen in the examples above these alleles may encode slightly differnt versions of a protein giving rise to different phenotypic traits. A gene’s most common allele is call the wild type with rare or uncommon alleles called mutants. Natural selection and genetic drift determine the relative frequencies of the alleles in a population.
Single Nucleotide Polymorphism - SNP
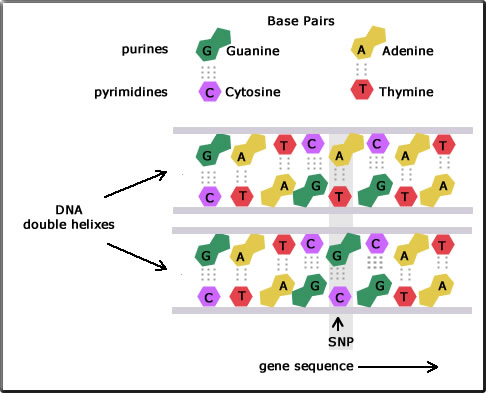
As has already been mentioned DNA replication is for the most part extremely accurate. However, a single base can be changed during DNA replication as is shown in the diagram. Here a single nucleotide - A, C, G, or T - in part of the DNA chain differs between two bees.
For example, the diagram shows two DNA fragments for the two individuals. The top sequences in the strands are shown to be: GATCACAT and GATCGCAT. Here a single nucleotide is seen to be different: A in the top fragment and G in the one below. This is called a point mutation. Because the nucleotide sequence along the gene is different, there are now two alleles. The stretch of the DNA where this change occurs may be within the coding region of the gene, or the non-coding region of the gene or in the intergenic region.
When more than 1% of a population carry this change then it is called an SNP (pronounced snip) and it becomes a genetic marker for that population.
For example, the diagram shows two DNA fragments for the two individuals. The top sequences in the strands are shown to be: GATCACAT and GATCGCAT. Here a single nucleotide is seen to be different: A in the top fragment and G in the one below. This is called a point mutation. Because the nucleotide sequence along the gene is different, there are now two alleles. The stretch of the DNA where this change occurs may be within the coding region of the gene, or the non-coding region of the gene or in the intergenic region.
When more than 1% of a population carry this change then it is called an SNP (pronounced snip) and it becomes a genetic marker for that population.